
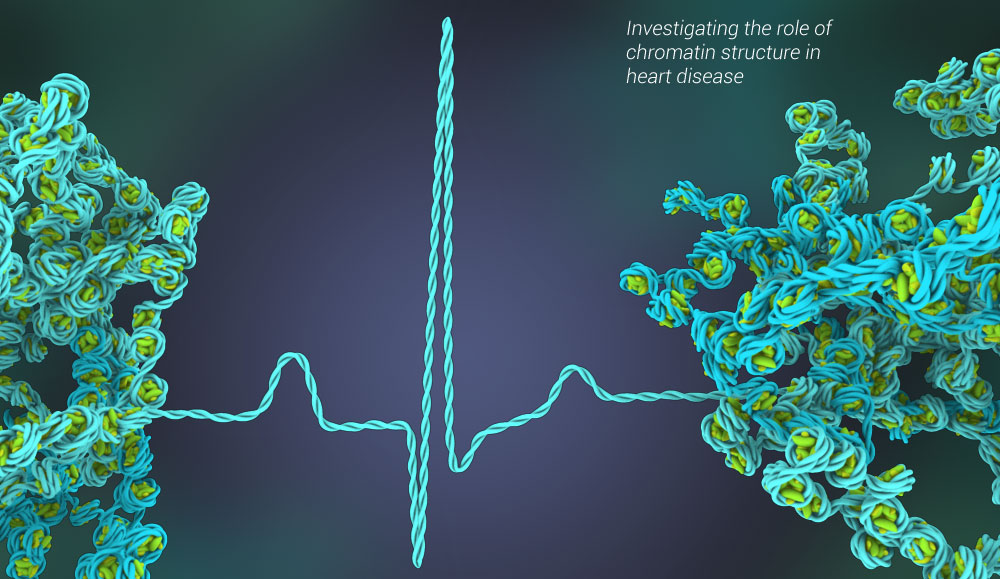
The Franklin Laboratory’s research is focused on the nucleus, DNA is wrapped around histone proteins (i.e. nucleosomes). This combination of DNA, histones and other proteins is referred to as chromatin. Both the DNA and proteins in this complex are susceptible to modifications which can alter the chromatin structure and consequently influence gene transcription. Changes in gene transcription can influence cell fate and physiology and have been shown to be altered in diseases such as heart failure. Because more people die from heart disease than any other pathology, the Franklin lab is interested in identifying the epigenetic factors that regulate gene transcription in the heart during disease progression. To do this, the lab uses a combination of proteomics, mass spectrometry, biochemistry and molecular biology to elucidate the role of histone isoforms, post-translational modifications and other chromatin binding proteins on chromatin structure and gene accessibility. Additionally, the Franklin lab also determines how these factors contribute to the regulation of heart morphology and physiology.
Featured Publications
Transcriptional regulation by methyltransferases and their role in the heart: highlighting novel emerging functionality | we focused our analysis on the largest subgroup, that of protein methyltransferases, and present a newly emerging phenomenon in which 16 of these enzymes have been shown to play dual roles in regulating transcription by maintaining the ability to both activate and repress transcription through methyltransferase-dependent or -independent mechanisms. | Szulik MW, Davis K, Bakhtina A, Azarcon P, Bia R, Horiuchi E, Franklin S. (2020) Transcriptional regulation by methyltransferases and their role in the heart: highlighting novel emerging functionality | |
Reductive Stress Causes Pathological Cardiac Remodeling and Diastolic Dysfunction | Formation of new neurons (neurogenesis), maturation (i.e. dendritic and axonal development) and integration into the entire neuronal network are central for gaining the functional plasticity. While there are limited therapeutic options currently available for neurodegenerative diseases, healing chronically injured neurons is still challenging | Shanmugam G, Wang D, Gounder SS, Fernandes J, Litovsky SH, Whitehead K, Radhakrishnan RK, Franklin S, Hoidal J, Kensler TW, Dell’Italia L, Darley-Usmar V, Abel ED, Jones DP, Ping P. Namakkal Soorappan R. (2020) Reductive Stress Causes Pathological Cardiac Remodeling and Diastolic Dysfunction. Antioxidants and Redox Signaling. | |
Molecular architecture of the Bardet-Biedl syndrome protein 2-7-9 subcomplex | Bardet-Biedl syndrome (BBS) is a genetic disorder characterized by malfunctions in primary cilia resulting from mutations that disrupt the function of the BBSome, an 8-subunit complex that plays an important role in protein transport in primary cilia. To better understand the molecular basis of BBS, here we used an integrative structural modeling approach consisting of EM and chemical cross-linking coupled with MS analyses, to analyze the structure of a BBSome 2-7-9 subcomplex consisting of three homologous BBS proteins, BBS2, BBS7, and BBS9. | Ludlam WG, Aoba T, Cuéllar J, Bueno-Carrasco MT, Makaju A, Moody JD, Franklin S, Valpuesta JM, Willardson BM. (2019) Molecular architecture of the Bardet-Biedl Syndrome protein core complex. |
The Franklin Lab Research Team
SARAH FRANKLIN, PH.D | ||
Katie Davis | Marta Szulik | Emilee Horiuchi |
Sam Hickenlooper | Magnus Creed | Ryan Bia |
Kylie Beach Graduate Research Assistant |













