


Personalized (or Precision) medicine is a rapidly advancing field of healthcare that is based in an individual’s unique genetic, environmental and clinical background. The principal goal is to identify genetic, genomic, environmental and clinical information that a physician can use to predict a patient’s susceptibility to a disease, the clinical course and the response to specific therapy. In order for precision medicine to realize its goals of improving healthcare delivery, a fundamental understanding of the genomic basis for disease susceptibility is required. The objective of our research program is to understand the genomic determinants of susceptibility to cardiac arrhythmia disorders and congenital heart disease with the goal of improving clinical management, risk stratification and clinical outcomes.
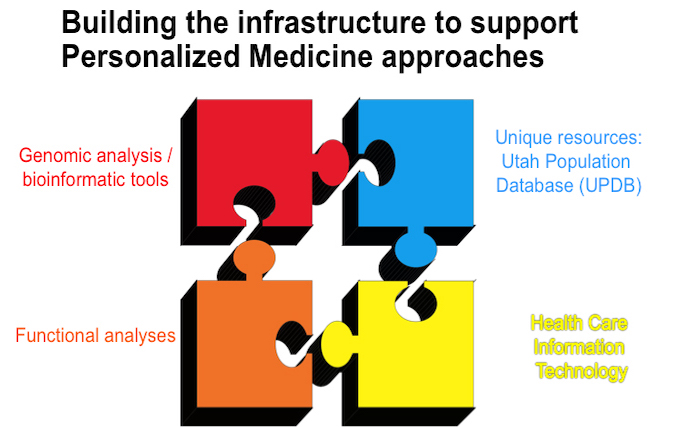
Utah is uniquely poised to address the challenges of precision cardiovascular medicine as we have the building blocks of a successful precision medicine program: state-of-art bioinformatic tools, the Utah Population Database, health care information technology systems and robust functional assays.

USTAR Center for Genetic Discovery (UCGD). In 2014, the State of Utah Science Technology and Research (USTAR) initiative invested $3.25 million, with a matching commitment of $3.25 million from the University of Utah Healthcare System, to establish the USTAR Center for Genetic Discovery. From DNA to diagnosis, the mission of the UCGD is to integrate patient genome information into health care. The UCGD develops tools for genome interpretation, disease discovery and big data management for research and clinical applications. The UCGD serves as the “computational engine” for several large-scale genomic medicine projects, including the Utah Genome Project and the Heritage 1K Project (a specific UGP project to sequence and analyze the whole genomes of 1,000 Utahans affected with conditions of interest). Core UCGD laboratories include the research groups of Mark Yandell, Ph.D., an internationally recognized leader in software development for comparative genomics and co-developer of the first probabilistic algorithm for sequence variant discovery; Gabor Marth, D.Sc., a computational biologist who has made significant contributions to national and international genome sequencing and interpretation projects including the Human Genome Project, the Cancer Genome Atlas Project, and the 1000 Genomes Project; and Aaron Quinlan, Ph.D., expert in cancer genome analysis and data integration tools and developer of BEDtools, a widely used software package for manipulating genomic data. Additional faculty associated with the UCGD are Lynn Jorde, executive director of the Utah Genome Project and lead author of the textbook Medical Genetics, and Karen Eilbeck, an expert in ontology development for biomedical knowledge representation.
Utah Population Database (UPDB). The UPDB is a rich source of information for genetic, epidemiological, demographic, and public health studies. The UPDB is the largest genealogical database in the world, housing genealogical records of more than 12 million individuals; these genealogical records are linked to medical records and Utah Department of Health vital statistics data. Through the identification and analysis of high-risk pedigrees, UPDB-based research defined the genetic risk factors for complex traits ranging from breast, colon and skin cancers to birth defects and congenital heart disease. It is important to note that the Utah population is genetically similar to other populations of European ancestry (validated by the Human Genome and HapMap Projects), and therefore, our data is generalizable to other populations of Northern and Western European ancestry. Finally, the use of high-risk pedigrees offers distinct advantages for gene discovery compared to isolated case cohorts or trios by eliminating potential variants that do not segregate with the phenotype to enhance the likelihood of identifying a high-penetrance mutation. Our research projects leverage the UPDB resources in order to facilitate the identification of disease-causing alleles and then validate these alleles in the larger research program datasets.
Functional Assays

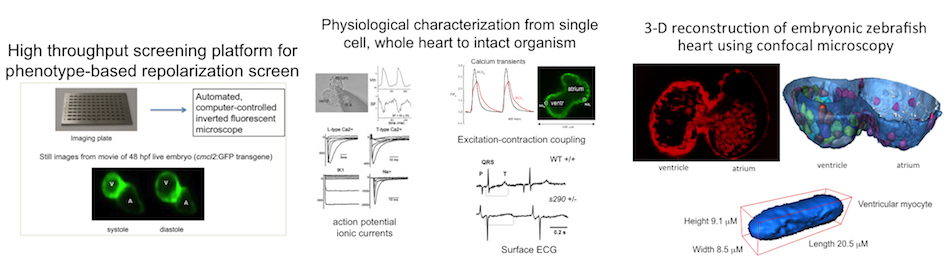
Zebrafish Model Organism. The zebrafish is especially relevant to the study of human arrhythmia as fundamental electrical properties are similar to higher mammals. Due to the large number of genetically identical offspring, the optical translucency of embryos and the availability of cardiac-specific transgenic fluorescent reporters, zebrafish are ideally suited for high-throughput phenotype- or physiology-based screens. Recently, we combined our high-throughput experience with our established LQTS zebrafish model to design a functional assay that distinguishes between pathogenic and benign human KCNH2 variants, highlighting that our research team has both the expertise and infrastructure to carry out high-throughput functional analyses in zebrafish. Moreover, we designed a system for high-throughput physiological characterization of candidate cardiovascular disease-associated genes, using an antisense morpholino-based (and more recently CRISPR/Cas9) knockdown approach and an automated, computer-controlled system. Our functional tool kit ranges from simple physiological read-outs (heart rate, rhythm, contractility, chamber size, morphology) to more complex physiological and morphologic measures (action potentials, calcium transients and 3D reconstructions). These assays allow us to test the functional consequences of putative cardiovascular disease-causing genes in this powerful vertebrate model organism.
Induced pluripotent stem cell derived cardiomyocytes (iPSC-CMs)
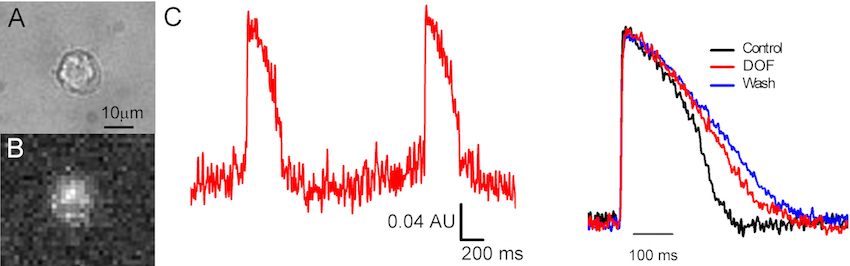
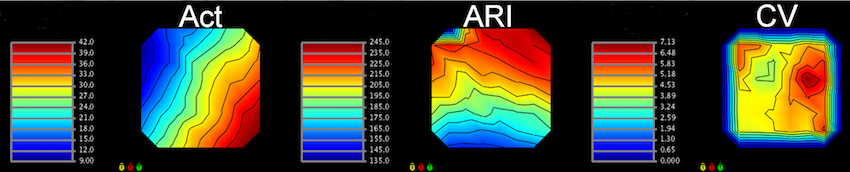
The reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) has revolutionized the fields of biology and medicine, providing new avenues for research and regenerative medicine applications. IPSCs are typically generated from somatic cells by overexpression of a set of pluripotency-specific transcription factors. We established a robust human iPSC-CM model system for the study of arrhythmia disorders. We were the first lab to characterize the electrophysiological and pharmacological properties of CMs using human peripheral blood mononuclear cell as a somatic cell source for IPSCs. A major impediment to the widespread use of human iPSC-CM assays is the low throughput of the currently available electrophysiological tools. We have overcome this hurdle by using a near-infrared fluorescent voltage-sensitive dye (di-4-ANBDQBS) to increase throughput >10-fold over traditional patch clamp technique. This system allows for the moderate-throughput characterization of the electrophysiological properties of patient- and disease-specific iPSC-CM.